Professional Manufacturer of Biomagnetic Beads
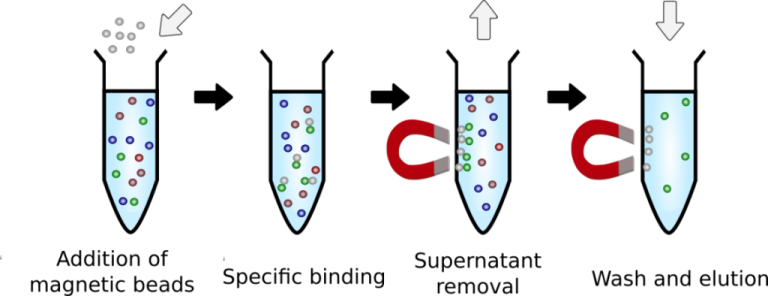
Solid phase reversible immobilization (SPRI) techniques have been used for DNA isolation for the past 20 years. In it, superparamagnetic carboxylated magnetic beads are used to selectively precipitate DNA molecules of different sizes under conditions of polyethylene glycol (PEG) percentage and salt concentration, and the products obtained can be used to prepare NGS sample libraries. One of the key points of the method is the determination of the critical PEG value since this is the most important parameter for the selective separation of DNA fragments.
Earlier studies have found that selective isolation of DNA fragments is associated with a conformational shift in DNA morphology from coiled-coil to globular. Previously, researchers have used a variety of methods to determine the critical PEG concentration that induces the transition of DNA molecules from coiled to spherical, such as sedimentation, fluorescence imaging, viscosity, UV/Vis centrifugation, and dynamic light scattering (DLS), and circular dichroism. However, these methods did not apply to DNA solutions containing magnetic beads because the globular DNA molecules would precipitate onto solid surfaces. Therefore, later researchers have used gel electrophoresis to determine the critical PEG concentration for DNA fragments of a specific size, however, this method has several disadvantages, such as the relatively long electrophoresis time and less accurate results for critical values.
Therefore, in 2013, He Zhangyong et al [1] used to study the effect of different concentrations of polyethylene glycol (PEG) on the recovery of DNA fragments in the presence of 1 μm carboxylated magnetic beads at a fixed concentration of sodium chloride (NaCl). They also fitted their concentrations to the PEG concentration by a logistic function and determined the critical PEG value from the fitted logistic function. On this basis, they obtained the slope function of the recovery equation by deriving its first-order derivatives and used the slope function based on these parameters to build a DNA fragment recovery spectrum which was able to determine the concentration of NaCl and PEG for the separation of different fragments [2]. In addition to this, they found in their study that pH has no effect on the effectiveness of their system and that the molecular weight size of PEG is inversely proportional to the size of the concentration required.
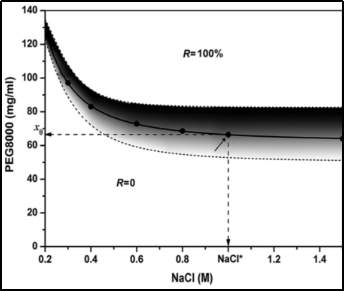
Whereas earlier E. Froehlich et al [3] investigated the binding of DNA to polyethylene glycols (PEG) of different sizes and compositions (e.g., PEG 3350, PEG 6000, and MPEG-anthracene) in salt-ionic solutions. Structural analysis showed that PEG 3350 and PEG 6000 formed moderate complexes, whereas the mPEG-anthracene-DNA adducts had weak interactions with hydrophilic and hydrophobic groups in contact. The stability of the complexes formed was in the order of PEG 6000 > PEG 3350 > mPEG-anthracene, with almost one PEG molecule bound per DNA. No conformational changes were observed for B-DNA, whereas at higher PEG concentrations the DNA condensed and formed particles.
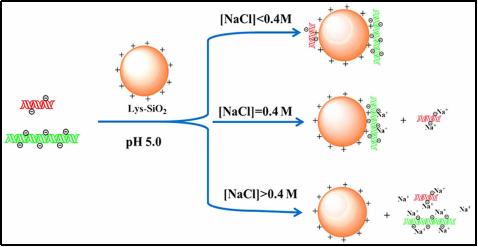
In addition, Lingling Liu et al[2] demonstrated that lysine silica particles could be used for size-selective separation of binary and ternary DNA mixtures by adjusting the electrostatic interactions between DNA fragments and the surface of lysine-functionalized silica particles according to the ambient pH, in particular, the ionic strength. The charge of the DNA fragments, and the electrostatic interactions between them and the surface of oppositely charged silica particles, was determined by the pH of the environment. The charge of DNA fragments and their electrostatic interactions with the surface of the oppositely charged silica particles are closely related to the size of the DNA fragments, and the mechanism of selective separation of DNA fragments using Lys-SiO2 particles is shown in Figure 1.
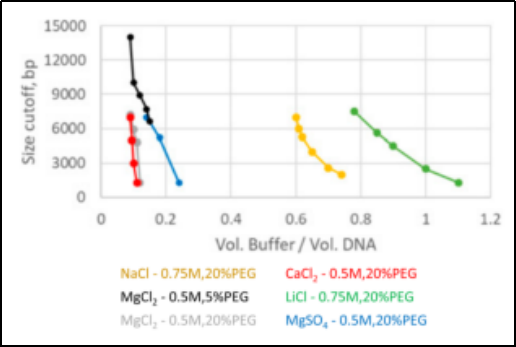
In 2020 Alexei et al [4] shifted the interval of selective isolation of DNA fragments of 150-800 bp to 1.5-7 Kbp by adjusting the concentration of NaCl in the magnetic bead buffer. When replacing NaCl with other cations and lowering the concentration of polyethyleneglycol (PEG) 8000, the selectively separated DNA fragment intervals varied, with LiCl being the most effective here.
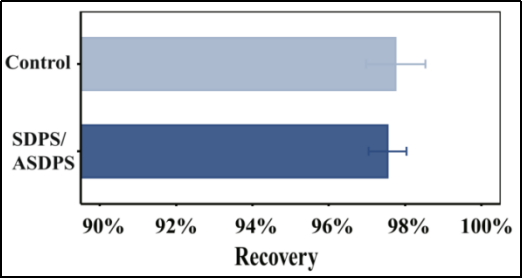
2023 Danli et al [5] tested the maximized recovery of critical concentrations of polyethylene glycol (PEG) 8000 and 300 nm carboxylated magnetic beads, determining a magnetic bead buffer composition for carboxylated magnetic beads of 20% PEG 8000, 2M NaCl, and 16.3 mM MgCl2, along with 1.25 mg/ml magnetic beads. They then tested the protocol for DNA recovery comparable to a control Beckman (AMPure XP magnetic beads).
Reserve buffers are not universal due to the different parameters of different types of magnetic beads. However, any researcher can further optimize the magnetic bead suspension buffer based on known data results for efficient recovery or for different experimental purposes. From standard commercial DNA purification methods to adjusting SPRI bead concentrations and optimal bead suspension buffers, this can provide researchers with a great deal of flexibility and convenience for different DNA manipulation conditions. In addition, different DNA purification strategies have great potential for application in the biological and medical fields.
Reference:
[1] HE Z, ZHU Y, GU H. A new method for the determination of critical polyethylene glycol concentration for selective precipitation of DNA fragments[J/OL]. Applied Microbiology and Biotechnology, 2013, 97(20): 9175-9183. DOI:10.1007/s00253-013-5195-0.
[2] HE Z, XU H, XIONG M, et al. Size‐selective DNA separation: Recovery spectra help determine the sodium chloride (NaCl) and polyethylene glycol (PEG) concentrations required[J/OL]. Biotechnology Journal, 2014, 9(10): 1241-1249. DOI:10.1002/biot.201400234.
[3] PEG and mPEG–Anthracene Induce DNA Condensation and Particle Formation | The Journal of Physical Chemistry B[EB/OL]. https://pubs.acs.org/doi/10.1021/jp205079u.
[4] STORTCHEVOI A, KAMELAMELA N, LEVINE S S. SPRI Beads-based Size Selection in the Range of 2-10kb[J/OL]. Journal of Biomolecular Techniques : JBT, 2020, 31(1): 7-10. DOI:10.7171/jbt.20-3101-002.
[5] LIU D, LI Q, LUO J, et al. An SPRI beads-based DNA purification strategy for flexibility and cost-effectiveness[J/OL]. BMC Genomics, 2023, 24(1): 125. DOI:10.1186/s12864-023-09211-w.
Supplier
Shanghai Lingjun Biotechnology Co., Ltd. was established in 2016 which is a professional manufacturer of biomagnetic materials and nucleic acid extraction reagents.
We have rich experience in nucleic acid extraction and purification, protein purification, cell separation, chemiluminescence, and other technical fields.
Our products are widely used in many fields, such as medical testing, genetic testing, university research, genetic breeding, and so on. We not only provide products but also can undertake OEM, ODM, and other needs. If you have a related need, please feel free to contact us at sales01@lingjunbio.com.